Article preview View full access options
Nature | Letter
Mitochondrial ROS regulate thermogenic energy expenditure and sulfenylation of UCP1
- Journal name:
- Nature
- Volume:
- 532,
- Pages:
- 112–116
- Date published:
- DOI:
- doi:10.1038/nature17399
- Received
- Accepted
- Published online
Brown and beige adipose tissues can dissipate chemical energy as heat through thermogenic respiration, which requires uncoupling protein 1 (UCP1)1, 2. Thermogenesis from these adipocytes can combat obesity and diabetes3, encouraging investigation of factors that control UCP1-dependent respiration in vivo. Here we show that acutely activated thermogenesis in brown adipose tissue is defined by a substantial increase in levels of mitochondrial reactive oxygen species (ROS). Remarkably, this process supports in vivo thermogenesis, as pharmacological depletion of mitochondrial ROS results in hypothermia upon cold exposure, and inhibits UCP1-dependent increases in whole-body energy expenditure. We further establish that thermogenic ROS alter the redox status of cysteine thiols in brown adipose tissue to drive increased respiration, and that Cys253 of UCP1 is a key target. UCP1 Cys253 is sulfenylated during thermogenesis, while mutation of this site desensitizes the purine-nucleotide-inhibited state of the carrier to adrenergic activation and uncoupling. These studies identify mitochondrial ROS induction in brown adipose tissue as a mechanism that supports UCP1-dependent thermogenesis and whole-body energy expenditure, which opens the way to improved therapeutic strategies for combating metabolic disorders.
Subject terms:
At a glance
Figures
-
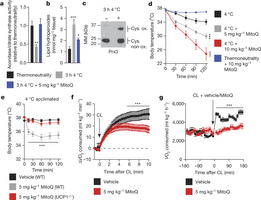
Figure 1: Increased BAT mitochondrial ROS levels support UCP1-dependent thermogenesis in vivo. a–c, Effect of acute cold exposure on in vivo BAT (a) mitochondrial superoxide-dependent inactivation of mitochondrial aconitase (n = 5; 5 mg kg−1 MitoQ n = 4), (b) lipid hydroperoxide content (n = 5), and (c) mitochondrial hydrogen-peroxide-dependent oxidation of Prx3. Oxidized Prx3 was assessed using the redox gel shift method described in Methods. MM, molecular mass. Cys. ox., cysteine oxidation. For uncropped scans see Supplementary Figs 1–3. d, Effect of i.p. MitoQ on core body temperature after acute cold exposure (n = 10). e, Effect of i.p. MitoQ on core body temperature of WT and UCP1−/− mice after 4 °C acclimation (n = 10; UCP1−/− n = 8). f, Oxygen consumed acutely before and after i.p. CL ± MitoQ (n = 5). g, Oxygen consumed 3 h before and after i.p. CL ± MitoQ (n = 8). Data are mean ± s.e.m. of at least four mouse replicates. *P < 0.05, ***P < 0.001 (two-tailed Student’s t-test for pairwise comparisons, one-way/two-way analysis of variance (ANOVA) for multiple comparisons involving one/two independent variables).
-
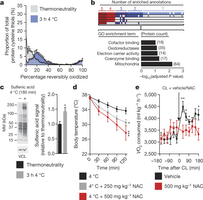
Figure 2: BAT mitochondrial ROS during thermogenesis drives oxidation of cellular and mitochondrial thiols. a, Distribution of percentage reversible oxidation status of BAT protein thiols ± acute cold exposure. b, Pathway analysis of BAT proteins containing cysteine residues sensitive to substantial oxidation (>10% shift in oxidation status) upon cold exposure. Top: proteins clustered according to shared GO enrichment terms. Bottom: significantly enriched pathways. c, Immunodetection of protein sulfenic acid levels in BAT ± acute cold exposure (n = 4) d, Effect of i.p. NAC on core body temperature after acute cold exposure (n = 8; 500 mg kg−1 NAC n = 7). e, Oxygen consumed 3 h before and after i.p. CL ± NAC (control n = 12; NAC n = 9). VCL, vinculin. Data are mean ± s.e.m. of at least four mouse replicates. *P < 0.05, **P < 0.01, ***P < 0.001 (two-tailed Student’s t-test for pairwise comparisons, one-way/two-way ANOVA for multiple comparisons involving one/two independent variables).
-
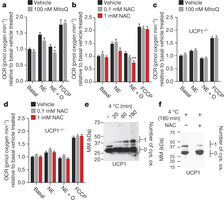
Figure 3: BAT mitochondrial ROS oxidatively modify a cysteine residue on UCP1 and support UCP1-dependent leak respiration. a, b, OCR of brown adipocytes ± noradrenaline (NE) stimulation + oligomycin to determine leak respiration ± (a) MitoQ (n = 10) or (b) NAC (n = 8; 0.1 mM NAC n = 7). c, d, OCR of primary UCP1−/− adipocytes ± noradrenaline stimulation + oligomycin ± (c) MitoQ (n = 10) or (d) NAC (n = 10). e, f, Cys-redox immunoblot of BAT UCP1 (e) after a time course of acute cold exposure, and (f) after acute cold exposure ± 500 mg kg−1 NAC. FCCP, carbonyl cyanide-p-trifluoromethoxyphenylhydrazone. Data are mean ± s.e.m. of at least seven cellular replicates. *P < 0.05, **P < 0.01 (two-tailed Student’s t-test for pairwise comparisons, one-way ANOVA for multiple comparisons).
-
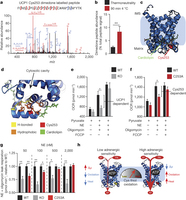
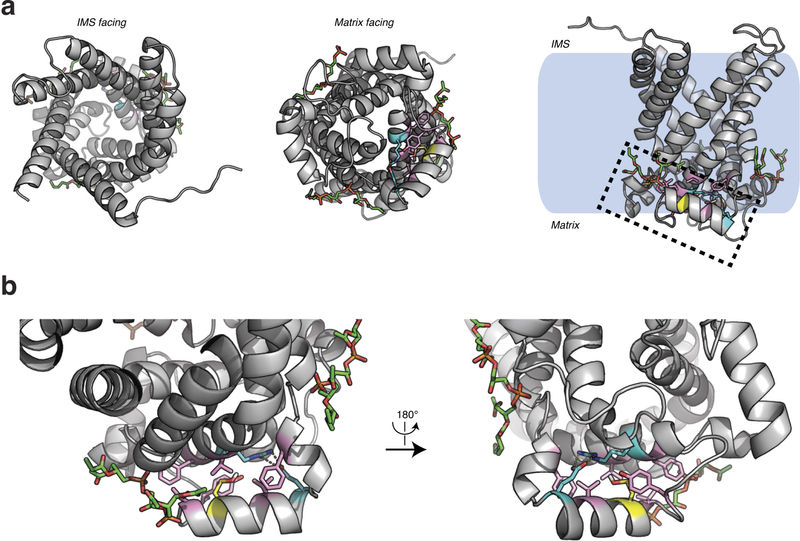
Figure 4: UCP1 Cys253 is sulfenylated during thermogenesis and sensitizes UCP1 to adrenergic activation. a, MS2 spectrum of dimedone-labelled UCP1 Cys253 peptide indicating sulfenylation of this site during thermogenesis. Fragment ions that span the dimedone-alkylated cysteine are highlighted in the peptide sequence. C#, dimedone-labelled cysteine; M*, oxidized methionine. b, Quantification of dimedone-labelled UCP1 Cys253 relative to the NEM-alkylated form (n = 5). c, d, Structure of (c) human UCP1 modelled on the AAC crystal structure and (d) Cys253 in a hydrophobic pocket between two matrix facing helices. IMS, intermembrane space. e, f, Basal, maximal, and UCP1-dependent OCR of UCP1−/− brown adipocytes ± transduction with (e) WT UCP1 (WT n = 7; UCP1−/− n = 6) or (f) UCP1 C253A (WT n = 17; C253A n = 19). KO, knockout. g, UCP1-dependent leak respiration after stimulation by various concentrations of noradrenaline (50 nM noradrenaline n = 9; 100 nM noradrenaline n = 7; 500 nM noradrenaline n = 8; 2,000 nM noradrenaline n = 6). Comparison of UCP1 WT and C253A indicates that degree of UCP1 inhibition by C253A is inversely correlated with noradrenaline concentration (n = 19; 100 nM WT noradrenaline; n = 17,500 nM noradrenaline n = 18; 2,000 nM noradrenaline n = 10). j, A model of sensitization of UCP1-mediated uncoupling by mitochondrial ROS. Data are mean ± s.e.m. of at least five mouse replicates or cell replicates for respirometry experiments. *P < 0.05, **P < 0.01 (two-tailed Student’s t-test for pairwise comparisons).
-
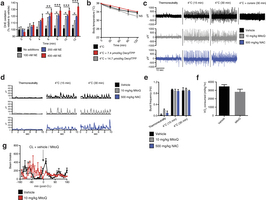
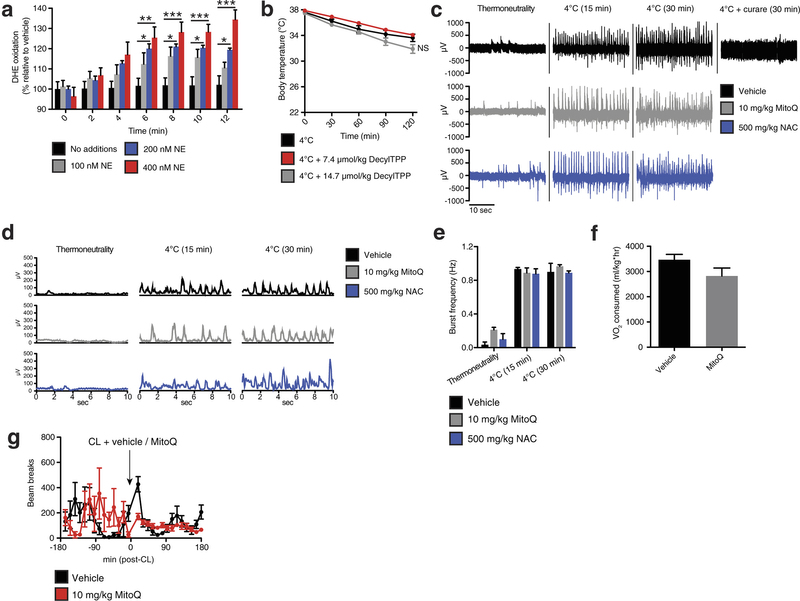
Extended Data Fig. 1: Assessing superoxide in brown adipocytes and effect of mitochondria-targeted compounds on shivering, body temperature, and movement. a, Noradrenaline stimulates superoxide-dependent oxidation of DHE in primary brown adipocytes (n = 5). b, Effect of i.p. decyl-TPP on core body temperature after acute cold exposure (n = 10). c, d, Representative (c) raw and (d) root mean square mouse EMG traces at thermoneutrality and after acute cold exposure ± MitoQ, NAC, or curare (0.3 mg kg−1). e, Quantification of muscle burst frequency as determined by EMG at thermoneutrality and after acute cold exposure ± MitoQ or NAC (n = 3). f, Absolute oxygen consumed immediately before acute CL treatment described in Fig. 1f (n = 5). g, Effect of i.p. CL ± MitoQ on movement as assessed by number of beam breaks (n = 8). NS, not significant. Data are mean ± s.e.m. of at least three replicates. *P < 0.05, **P < 0.01, ***P < 0.001 (two-tailed Student’s t-test for pairwise comparisons, one-way ANOVA for multiple comparisons, two-way ANOVA for multiple comparisons involving two independent variables).
-
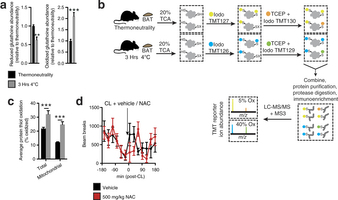
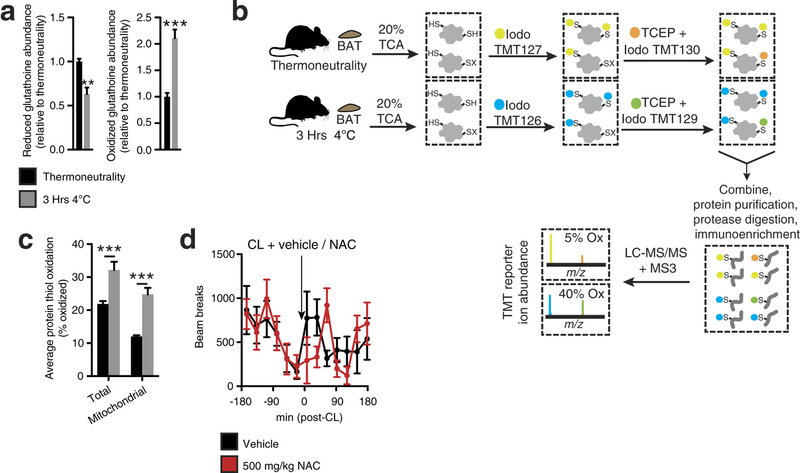
Extended Data Fig. 2: Assessing thiol redox status in vivo and the effect of NAC on movement. a, Mass spectrometric quantification of BAT-reduced and -oxidized glutathione at thermoneutrality and after acute cold exposure (n = 5). b, Scheme for quantitative assessment of protein thiol redox status by ratiometric labelling of BAT protein cysteine thiols. In vivo BAT thiol status is stabilized by protein precipitation in 20% TCA49. Unmodified cysteine thiols are labelled with ‘light’ iodoTMT tags (126, 127, 128). After removal of light iodoTMT, reversibly modified protein thiols are reduced with TCEP in the presence of ‘heavy’ iodoTMT (129, 130, 131). Samples are combined and subjected to trypsin digestion. Ratiometric assessment of iodoTMT labelled peptides provides a quantitative profile of overall protein cysteine thiol redox status. c, Average percentage oxidation status of total BAT and BAT mitochondrial protein thiols at thermoneutrality and after acute cold exposure (n = 3). d, Effect of i.p. NAC on movement as assessed by number of beam breaks (vehicle, n = 11; NAC, n = 7). Data are mean ± s.e.m. of at least three replicates. *P < 0.05, ***P < 0.001 (two-tailed Student’s t-test for pairwise comparisons, one-way ANOVA for multiple comparisons, two-way ANOVA for multiple comparisons involving two independent variables).
-
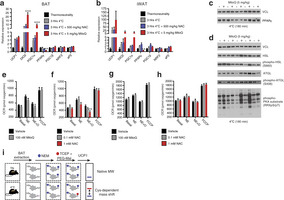
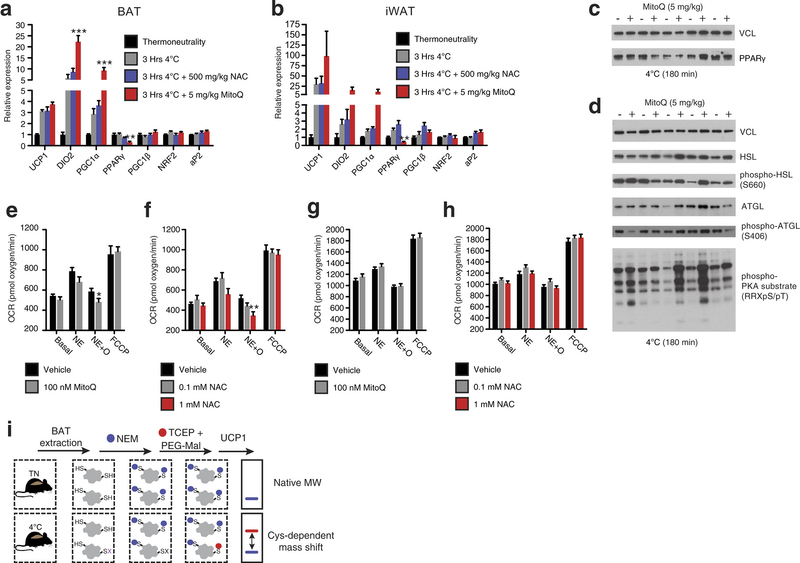
Extended Data Fig. 3: Assessing thermogenic gene expression, adrenergic response parameters, and strategy for determination of UCP1 cysteine thiol redox status. a, b, Quantitative polymerase chain reaction (qPCR) analysis of mRNA expression of selected (a) BAT and (b) inguinal white adipose tissue (iWAT) genes ± cold exposure, ± MitoQ or NAC (n = 5). c, d, Immunoblot analysis of (c) PPAR-γ protein expression levels and (d) lipolytic phosphorylation cascade activation in BAT after cold exposure ± MitoQ. e, f, Raw OCR of primary brown adipocytes under basal conditions and after noradrenaline stimulation + oligomycin to determine leak respiration ± (e) MitoQ (n = 10) or (f) NAC (vehicle and 1 mM NAC, n = 8; 0.1 mM NAC, n = 7). g, h, OCR of primary brown adipocytes lacking UCP1 under basal conditions and after noradrenaline stimulation + oligomycin ± (g) MitoQ (n = 10) and (h) NAC (n = 10). i, Cys-redox mass shift strategy. After in vivo interventions, mouse BAT is excised and unmodified protein thiols are labelled with NEM, after which reversibly oxidized thiols are chemically reduced and labelled with polyethyleneglycol maleimide (PEG-Mal) allowing for determination of cysteine oxidation status on UCP1. Data are mean ± s.e.m. of at least five replicates. *P < 0.05, **P < 0.01 (two-tailed Student’s t-test for pairwise comparisons, one-way ANOVA for multiple comparisons, two-way ANOVA for multiple comparisons involving two independent variables).
-
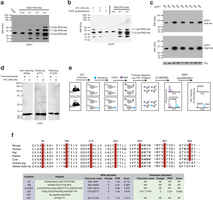
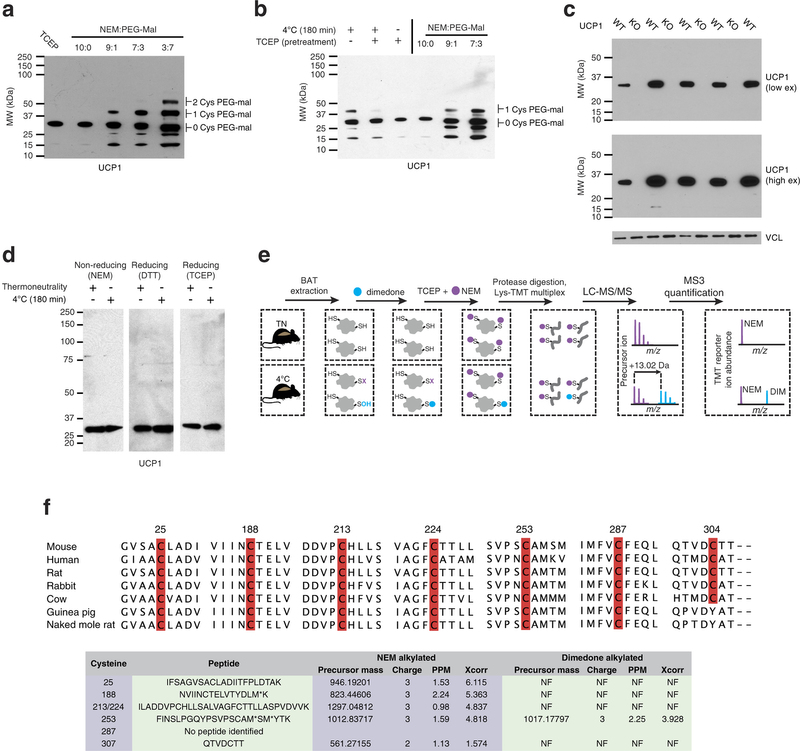
Extended Data Fig. 4: Assessing UCP1 reversible cysteine oxidation status in vivo by immunoblot and mass spectrometry. a, Calibration of UCP1 cysteine gel shift immunoblot. Calibration of cysteine-dependent shifts by incubation of BAT protein with TCEP and different ratios of NEM and PEG-Mal indicates that a single PEG-mal labelling event shifts UCP1 by ~10 kDa above the native molecular mass. b, Calibration of UCP1 cysteine oxidation status indicates that the gel shift observed upon cold exposure (lane 1) is cysteine dependent, as TCEP pretreatment results in a loss of the shift (lane 2). In addition, the cysteine-dependent mass shift is due to a single oxidation event as determined by including the calibrating markers (lanes 4–6). c, Calibration of specificity of UCP1 antibody in BAT. d, Reducing and non-reducing SDS–PAGE analysis of UCP1 to monitor cysteine-dependent and -independent inter-protein interactions during acute cold exposure. e, Scheme for identification of sulfenylated cysteines on UCP1 by dimedone labelling and mass spectrometry. After acute cold exposure, BAT protein thiols are differentially alkylated with dimedone to selectively label sulfenylated thiols and NEM to label non-sulfenylated thiols. Samples are subjected to trypsin digestion, followed by Lys-TMT labelling, and MS quantification of UCP1 cysteine containing peptides in their dimedone and NEM alkylated forms. Two technical points should be noted in this strategy when interpreting relative quantitation of NEM and dimedone-alkylated peptides. First, these differently alkylated peptides may not necessarily ionize with the same efficiency. Second, NEM is reported to react with sulfenic acids albeit less efficiently than with free thiols50, which should be factored in when considering the order of addition of dimedone/NEM and potential underestimation of sulfenylation status. f, Top: amino-acid sequence alignment of UCP1 proteins highlighting the candidate cysteine residues contained within the mouse protein and their level of conservation across various species. Bottom: summary of MS determination of UCP1 cysteine sulfenylation status. Six of seven UCP1 cysteines were identified, with all but one being identified exclusively in the unmodified (NEM-alkylated state). Cys253 is identified as dimedone labelled, indicating that it is a site for sulfenylation.
-
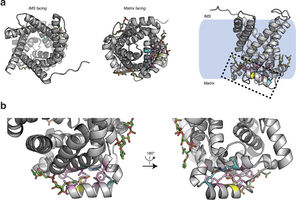
Extended Data Fig. 5: Structure of human UCP1 modelled on the AAC crystal structure including bound cardiolipin and sulfenylation of Cys253. a, Entire UCP1 modelled structure including bound cardiolipin (green), and Cys253 in the oxidized sulfenic acid form. b, UCP1 region containing Cys253 in the oxidized sulfenic acid form. Cys253 localizes to a hydrophobic pocket between two matrix facing helices. Hydrophobic residues (pink) surround the Cys253 thiol, and a hydrogen bond between Arg238 and Glu261 (aqua) is proximal. These residues that stabilize interaction between the matrix facing helices are probably important for stabilization of the purine bound ‘c-state’ of the carrier. Two separate cardiolipin (green) binding domains are localized proximal to Cys253 within the UCP1 modelled structure.
-
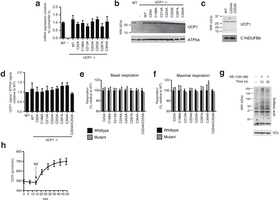
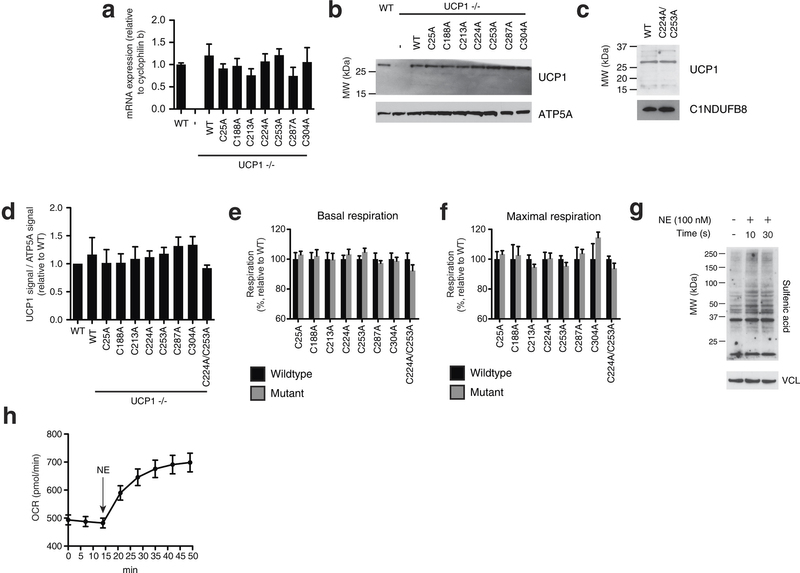
Extended Data Fig. 6: Assessing transduced UCP1 constructs, OCR, and sulfenylation in brown adipocytes. a, Quantitative PCR analysis of UCP1 mRNA in WT, and UCP1−/− brown adipocytes transduced with WT and cysteine null UCP1 mutants (n = 4). b, Immunoblot of UCP1 protein in WT and UCP1−/− brown adipocytes transduced with WT and cysteine null UCP1 mutants. c, Immunoblot analysis of UCP1 protein in UCP1−/− brown adipocytes transduced with C224A/C253A double mutant compared with WT. d, Densitometry analysis of transduced UCP1 forms relative to WT across separate transduction experiments (n = 4; C224A/C253A n = 3). e, f, Summary of basal (e) and maximal (f) OCR of primary brown adipocytes containing cysteine-null UCP1 mutants. Raw OCR values provided in Extended Data Fig. 7. g, Immunodetection of protein sulfenic acid levels in primary brown adipocytes in the seconds after treatment with 100 nM noradrenaline. h, Time course of brown adipocyte OCR after stimulation with 100 nM noradrenaline (n = 12). Data are mean ± s.e.m. of at least three replicates.
-
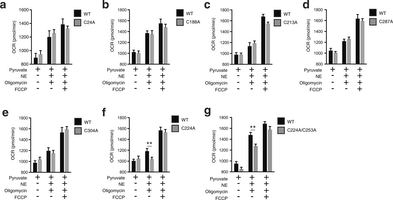
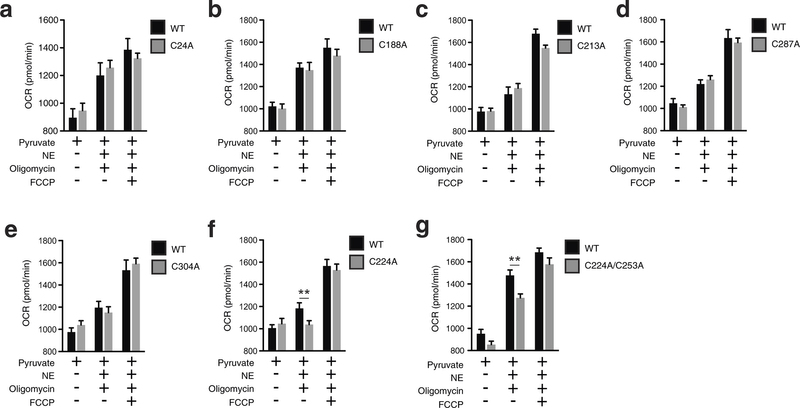
Extended Data Fig. 7: Assessing OCR under basal and FCCP-stimulated maximal respiration, and after noradrenaline stimulation + oligomycin in UCP1−/− primary brown adipocytes transduced with UCP1 cysteine null mutants. a–g, Raw basal, maximal, and UCP1-dependent OCR from representative experiments of WT and UCP1−/− brown adipocytes transduced with (a) UCP1 C24A (WT n = 11; C24A n = 12), (b) C188A (n = 7), (c) UCP1 C213A (n = 19), (d) UCP1 C287A (WT n = 9; C287A n = 10), (e) UCP1 C304A (WT n = 7; C304A n = 10), (f) UCP1 C224A (WT n = 9; C224A n = 10), (g) UCP1 C224A/C253A (n = 8). Data are mean ± s.e.m. of at least seven replicates. **P < 0.01 (two-tailed Student’s t-test for pairwise comparisons).
-
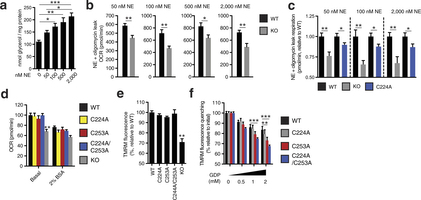
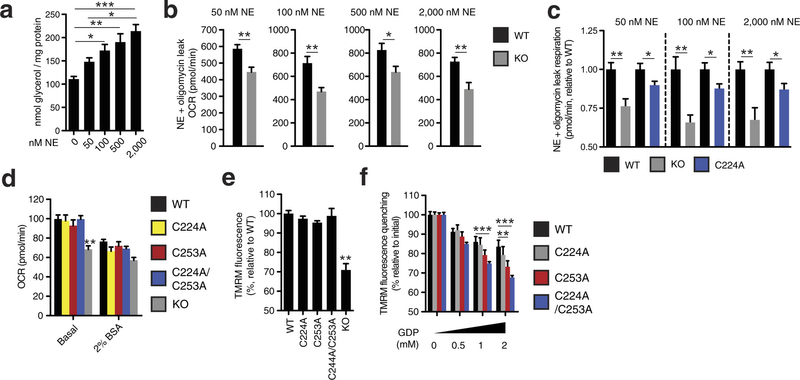
Extended Data Fig. 8: Assessing UCP1-dependent respiration and uncoupling following increasing degrees of adrenergic stimulus. a, Glycerol release from brown adipocytes as an index of adrenergic stimulus and lipolysis in response to increasing concentrations of noradrenaline (n = 4; 0 and 2,000 nM noradrenaline n = 6). b, Representative raw noradrenaline + oligomycin leak OCR values in WT and UCP1−/− brown adipocytes after stimulation with a range of noradrenaline concentrations indicates that UCP1-dependent leak respiration is consistently ~25–35% of total leak OCR (50 nM n = 9; 100 nM n = 7; 500 nM n = 8; 2,000 nM n = 6). c, Assessment of UCP1-dependent leak respiration after stimulation by various concentrations of noradrenaline + oligomycin. Comparison of WT and UCP1−/− OCR (replotted data from Fig. 4 for comparison) indicates that UCP1-dependent respiration is consistently ~25–35% of leak respiration. Comparison of UCP1 WT and C224A indicates that the degree of UCP1 inhibition by C224A is relatively stable across various noradrenaline concentrations (n = 8; C224A 100 nM noradrenaline n = 9, 2,000 nM noradrenaline n = 14). d, Plasma-membrane-permeabilized OCR of brown adipocytes ± endogenous fatty-acid depletion with BSA (WT n = 30; KO n = 20; C224A n = 9; C253A n = 8; C224A/C253A n = 10). e, Comparison of UCP1-dependent uncoupling absent purine nucleotide inhibition in plasma-membrane-permeabilized brown adipocytes (n = 6; C224A/C253A, KO n = 4). f, Comparison of UCP1-dependent uncoupling after titration of GDP in permeabilized adipocytes containing WT UCP1, UCP1 C224A, C253A, and C224A/C253A (n = 6; C224A/C253A n = 12). Data are mean ± s.e.m. of at least four replicates. *P < 0.05, **P < 0.01, ***P < 0.001 (two-tailed Student’s t-test for pairwise comparisons, one-way ANOVA for multiple comparisons).
Read the full article
Additional access options:
- British Library Document Supply Centre
- You can also request this document from your local library through inter-library loan services.